Ampure Xp Beads Size Selection Chart
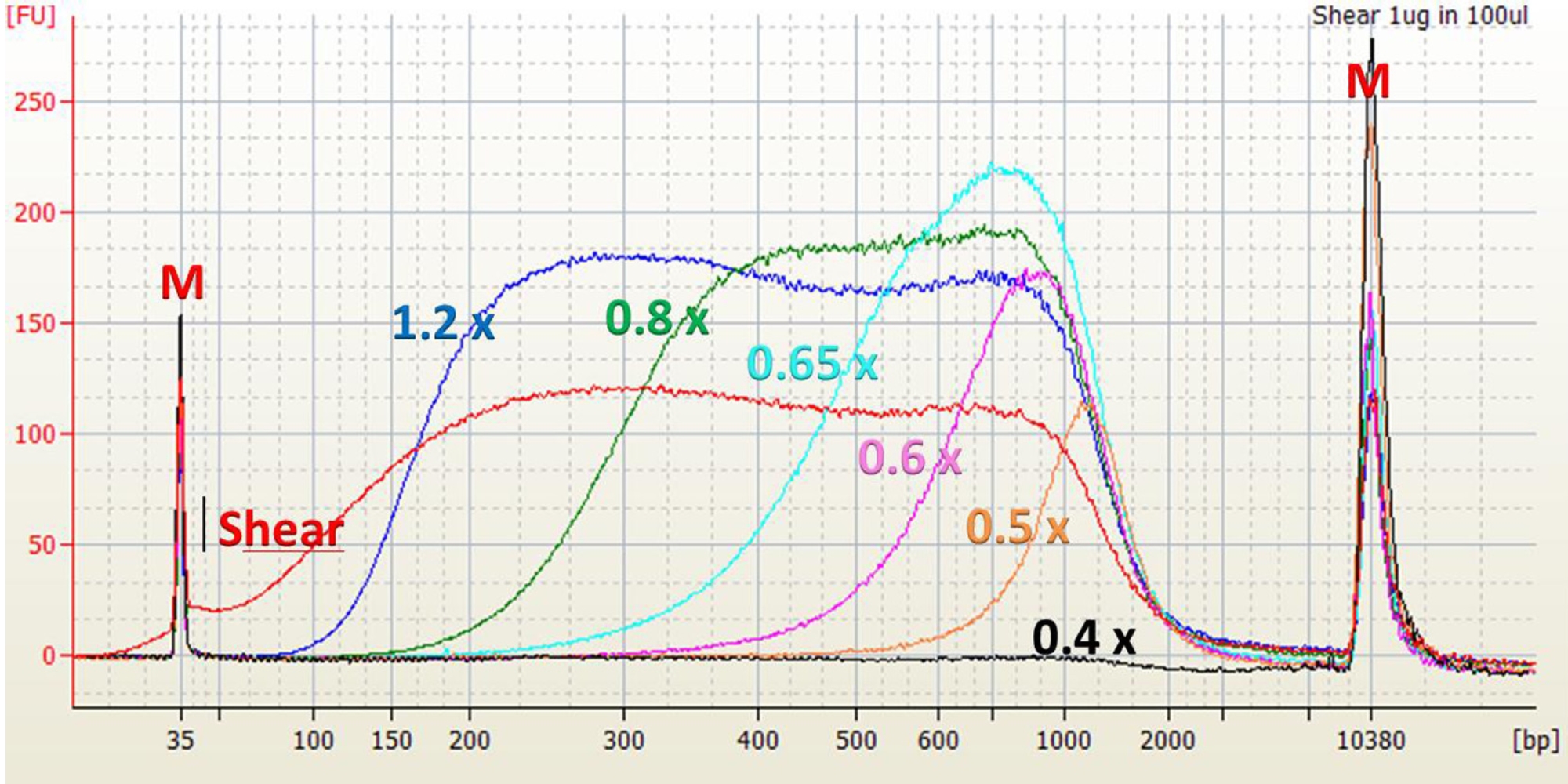
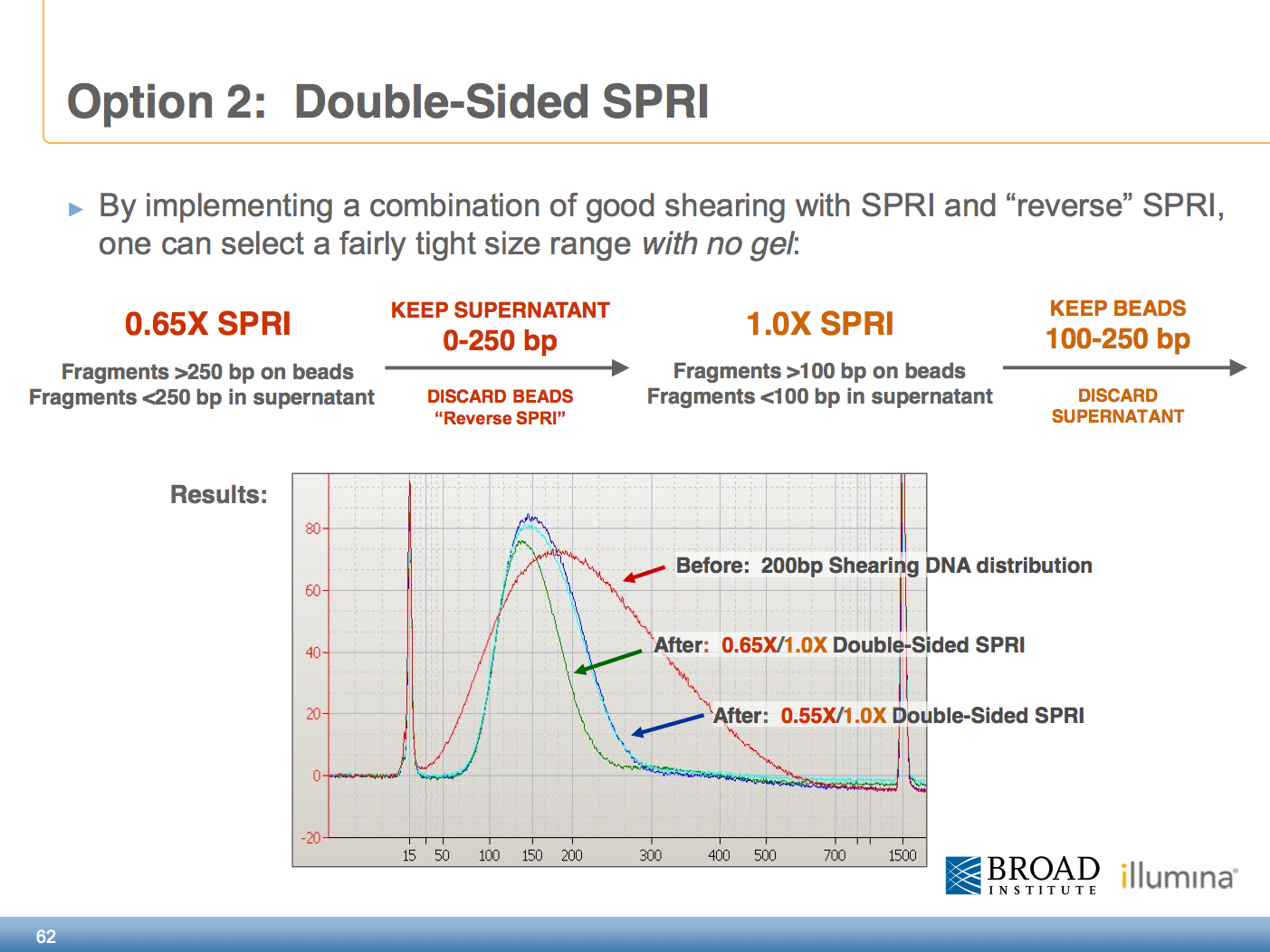
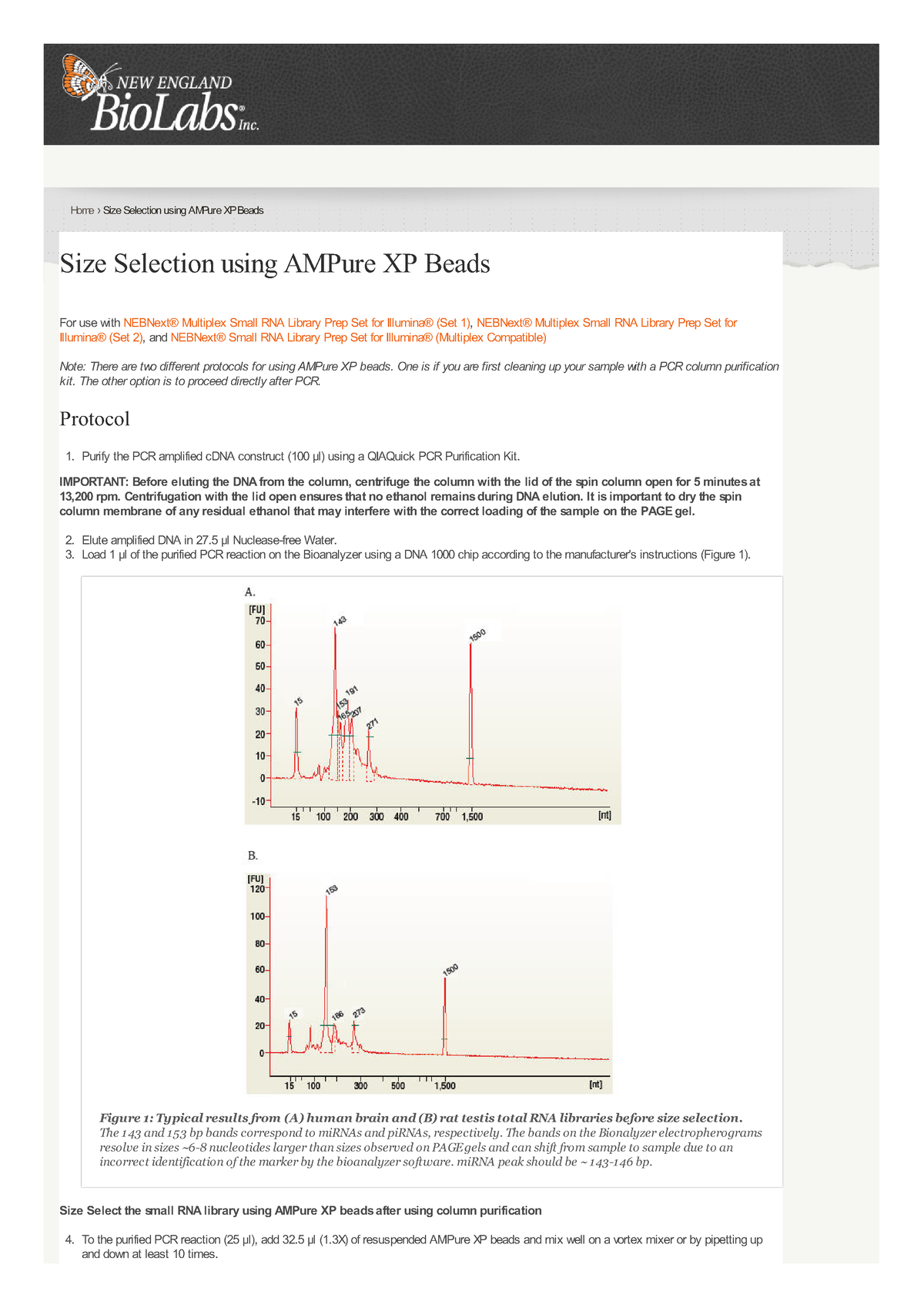
Ampure Xp Beads Size Selection Chart - Warm the ampure beads to room temperature and mix thoroughly before use. Excess primers, nucleotides, salts, and enzymes can. The following tables illustrate the number of pcr reactions th e agencourt ampure xp will. If you perform the qc check and your sample contains adaptor dimer (127 bp peak) or. It will be suitable to remove peaks >. Bead size selection is only recommended for samples showing no primer dimer and no adaptor dimer on bioanalyzer. Web we found that a 0.7x volume of custom spri beads (e.g. How do i perform size selection? Additionally, the dna concentration of the. Web the agencourt ampure xp can be used for pcr puri fication in 96 and 384 well format. Warm the ampure beads to room temperature and mix thoroughly before use. Web considerations for choosing an illumina dna pcr free library preparation workflow. Web the agencourt ampure xp can be used for pcr puri fication in 96 and 384 well format. Small molecular species such as free adapters and adapter. Web mix the total reaction volume by pipetting 10. Web we found that a 0.7x volume of custom spri beads (e.g. Web considerations for choosing an illumina dna pcr free library preparation workflow. Web standard ampure xp is used to remove dna fragments smaller than 100 bp. There are several different methods for performing size selection. Agencourt ampure xp beads) provides the best selection for dna fragments >2 kb,. Small molecular species such as free adapters and adapter. Web size select the small rna library using ampure xp beads after using column purification. Web considerations for choosing an illumina dna pcr free library preparation workflow. It is recommended to choose the appropriate method based on the qc check of the library. Web mix the total reaction volume by pipetting. Web we found that a 0.7x volume of custom spri beads (e.g. It will be suitable to remove peaks >. How do i perform size selection? There are several different methods for performing size selection. Additionally, the dna concentration of the. Additionally, the dna concentration of the. Web size select the small rna library using ampure xp beads after using column purification. This cut‐off can be increased or decreased (e.g., up to 300 bp, down to 50 bp) by isolating the beads. Web mix the total reaction volume by pipetting 10 times and incubate at rt for 1 minute or vortex. Agencourt ampure xp beads) provides the best selection for dna fragments >2 kb, and improves the median read. Although size selection protocols developed by other organizations do exist, we cannot guarantee performance or support this application. There are several different methods for performing size selection. It will be suitable to remove peaks >. It is recommended to choose the appropriate. Additionally, the dna concentration of the. Converting ng/µl to nm when calculating dsdna library concentration. Web agencourt ampure xp utilizes an optimized buffer to selectively bind dna fragments 100 bp and larger to paramagnetic beads. We suggest using the spriselect reagent , as it is developed. Agencourt ampure xp beads) provides the best selection for dna fragments >2 kb, and. Although size selection protocols developed by other organizations do exist, we cannot guarantee performance or support this application. Additionally, the dna concentration of the. Web can i perform size selection with the ampure xp reagent? Warm the ampure beads to room temperature and mix thoroughly before use. Excess primers, nucleotides, salts, and enzymes can. It is recommended to choose the appropriate method based on the qc check of the library. How do i perform size selection? It will be suitable to remove peaks >. Web we found that a 0.7x volume of custom spri beads (e.g. Web can i perform size selection with the ampure xp reagent? Web can i perform size selection with the ampure xp reagent? This cut‐off can be increased or decreased (e.g., up to 300 bp, down to 50 bp) by isolating the beads. Web size select the small rna library using ampure xp beads after using column purification. It allows you to perform. Web considerations for choosing an illumina dna pcr free. Web size select the small rna library using ampure xp beads after using column purification. Bead size selection is only recommended for samples showing no primer dimer and no adaptor dimer on bioanalyzer. We suggest using the spriselect reagent , as it is developed. Web standard ampure xp is used to remove dna fragments smaller than 100 bp. If you perform the qc check and your sample contains adaptor dimer (127 bp peak) or. Although size selection protocols developed by other organizations do exist, we cannot guarantee performance or support this application. To the purified pcr reaction (25 μl), add 32.5 μl (1.3x) of resuspended ampure xp. There are several different methods for performing size selection. Web considerations for choosing an illumina dna pcr free library preparation workflow. The following tables illustrate the number of pcr reactions th e agencourt ampure xp will. It is recommended to choose the appropriate method based on the qc check of the library. This cut‐off can be increased or decreased (e.g., up to 300 bp, down to 50 bp) by isolating the beads. Web agencourt ampure xp utilizes an optimized buffer to selectively bind dna fragments 100 bp and larger to paramagnetic beads. Small molecular species such as free adapters and adapter. Web size selection using ampure xp beads does not remove small fragments. Additionally, the dna concentration of the.
Ampure Beads Size Selection Chart
Ampure Xp Beads Size Selection Chart

Ampure XP Labplan

Ampure XP Labplan
How do SPRI beads work? Enseqlopedia

Optimising library size selection for iCLIP2 SampletoProNex bead

AMPure XP Beads Cleanup and Size Selection
Size Selection using AMPure XP Beads Accounting Studocu

Möglichkeiten mit AMPure XP Beckman Coulter

Bead Size Selection and Ratio Beckman Coulter
It Allows You To Perform.
Web We Found That A 0.7X Volume Of Custom Spri Beads (E.g.
It Will Be Suitable To Remove Peaks >.
Agencourt Ampure Xp Beads) Provides The Best Selection For Dna Fragments >2 Kb, And Improves The Median Read.
Related Post: